Cellular reprogramming is a promising new strategy for a variety of medical applications. One of many new and intriguing areas of biomedical engineering research, cellular reprogramming is a process where an adult cell is flipped from one type directly into another.
How can this be used for medical applications? Cellular reprogramming is being investigated to study and treat a variety of medical conditions such as cancer and sickle cell anemia, as well as for the regeneration of tissues lost due to injury or disease. In these cases, surrounding support cells provide a convenient and inexhaustible source for reprogramming. For example, CAR T cells are a promising new cancer therapy where a patient’s T cells are reprogrammed to target cancer cells. Another example is reprogramming scar tissue surrounding spinal cord injury into neural cells to restore neural circuitry and recover motor function. Reprogramming is also used for developing human disease models. Advances in tissue culture now allow for the in vitro spontaneous self-organization of complete organ-like structures from reprogrammed stem cells, allowing researchers to study the pathology and development of diseases in human models and test new treatments.
In this blog I will share some unpublished experimental results from a reprogramming project at the Willerth Lab at the University of Victoria where I conduct research.
Before cellular reprogramming technology can be understood a brief reminder of the flow of information within a cell is in order. All cells keep a master “blueprint” of the genetic code of the entire organism they belong to, known as the genome. In the cell genetic information flows from the genome, which is tightly packed inside the cell’s nucleus, to the cytoplasm where translational machinery known as ribosomes process it into proteins, the functional machinery of the cell. This is referred to as gene “expression”, and it is a highly regulated process developed throughout evolution.
A cell’s development from an undetermined or stem cell state into an adult cell with a specialized function is the result of a hardwired cascade of on/off gene expression. This begins with the expression of a few genes, which in turn trigger the expression of a series of developmental genes, eventually ending in the stable expression of a signature set of genes specific to a particular tissue or organ. Once a cell has acquired an adult fate any early developmental genes or genes related to alternate fates are silenced to preserve the stability of the cell throughout its lifetime. The key to cellular reprogramming is the identification of a type of regulatory gene with the unique ability to bind to silenced genes and activate them, triggering a new program within an adult cell while simultaneously suppressing the previous one.
Reprogramming genes have been identified for several distinct cell fates such as pancreatic beta cells, neural cells, blood cells, heart cells, and perhaps most notably stem cells – the early undefined cells which possess the features of indefinite self-replication and the ability to spontaneously give rise to all other cell types – a discovery that was awarded the Nobel Prize in 2012. These genes were discovered through screening processes in which potential genes are inserted into a cell’s genome using modified viruses and over-expressed, followed by gene expression analysis and functional testing to identify reprogrammed cell fates. These modified viruses are typically lentiviruses which have had their ability to use a host cell for self-replication removed followed by replacement of their viral genome with a gene of interest. Unfortunately this method of reprogramming is currently too risky for medical use, mainly because the gene of interest is randomly inserted into the genome, potentially causing harmful mutations in the process.
A safer alternative is to target the expression of the genome through its regulatory mechanisms. DNA expression is regulated by its accessibility. In order to squeeze the entire human genome into the tiny nucleus of a cell it is efficiently packaged away in a supercoiled structure known as chromatin, in which DNA is looped around histone proteins and bundled together in compact octets. In fact, for genetic information to escape the nucleus for translation, chromatin has to first be loosened at the site of the histones by regulatory enzymes known as ‘writers’ and ‘erasers’. These enzymes add or remove acetyl or methyl modifications to the histone proteins, altering their conformations and thus unraveling the chromatin to allow access to the DNA code. In addition to regulation at the chromatin level there is regulation at the transcriptional level. Incredibly, single cell transcriptome analysis has shown that nearly the entire genome is transcribed followed by active quality control by a diverse range of small non-coding RNA strands which freely circulate the nucleus, binding to rogue RNA before they can reach the cytoplasm for translation. The screening of huge libraries of small molecules and non-coding RNAs has led to the discovery of highly efficient “cocktails” of small molecules and small RNAs for cellular reprogramming.
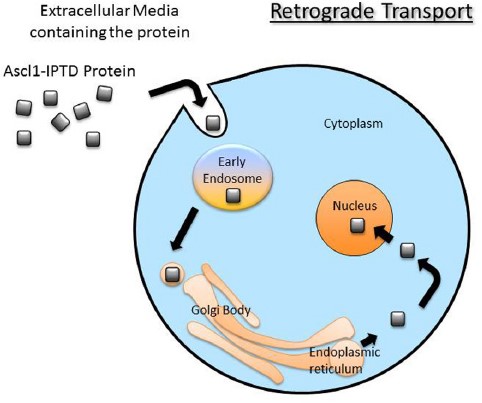
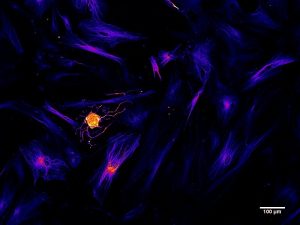
Another avenue being explored is functionalizing genes of interest with intracellular delivery sequences. I will use an example from my research in collaboration with iProgen Biotech to illustrate this type of reprogramming. iProgen Biotech has designed a single polypeptide chain combining ASCL1, a reprogramming gene for neural cells, with an amino acid secretion sequence directing it through the cell via a transport pathway known as “retrograde” transport. It enters the cell through selective channels and travels through the Golgi Apparatus (an organelle whose role is to sort and process proteins) to the nucleus where it initiates reprogramming by binding to silenced genes responsible for neural cell development. In the Willerth Lab at the University of Victoria we tested this functionalized ASCL1 protein on human brain astrocytes, a type of support cell that grows in abundance in the central nervous system. After 2 weeks of introducing the protein to the extracellular environment of our astrocytes in vitro we saw the appearance of neural cells, identified by fluorescently tagging the neural cytoskeletal protein beta tubulin (TUJ1).
As I noted at the beginning of this article, cellular reprogramming is being investigated to study and treat a variety of medical conditions. I would enjoy hearing from readers about their medical applications and experiments involving the technology.
Meghan Robinson is a part-time StarFish Medical BioMedical Engineering Lab Technician. This is her first StarFish Medical blog. In addition to her work with StarFish Medical, Meghan researches cell reprogramming under Dr. Stephanie Willerth at the University of Victoria and is the lead author on a number of peer reviewed publications in this field.
Images: University of Victoria