When selecting a method for developing an assay, whether it is for detecting bacteria or simulating fluid, the approach all depends on the application and your target market. For example, Escherichia coli (or E. coli as it is more commonly known) can be found everywhere, from lettuce sold at a supermarket to your kids’ diaper. There are many strains of E. coli, but only a few that we really need to be concerned about.
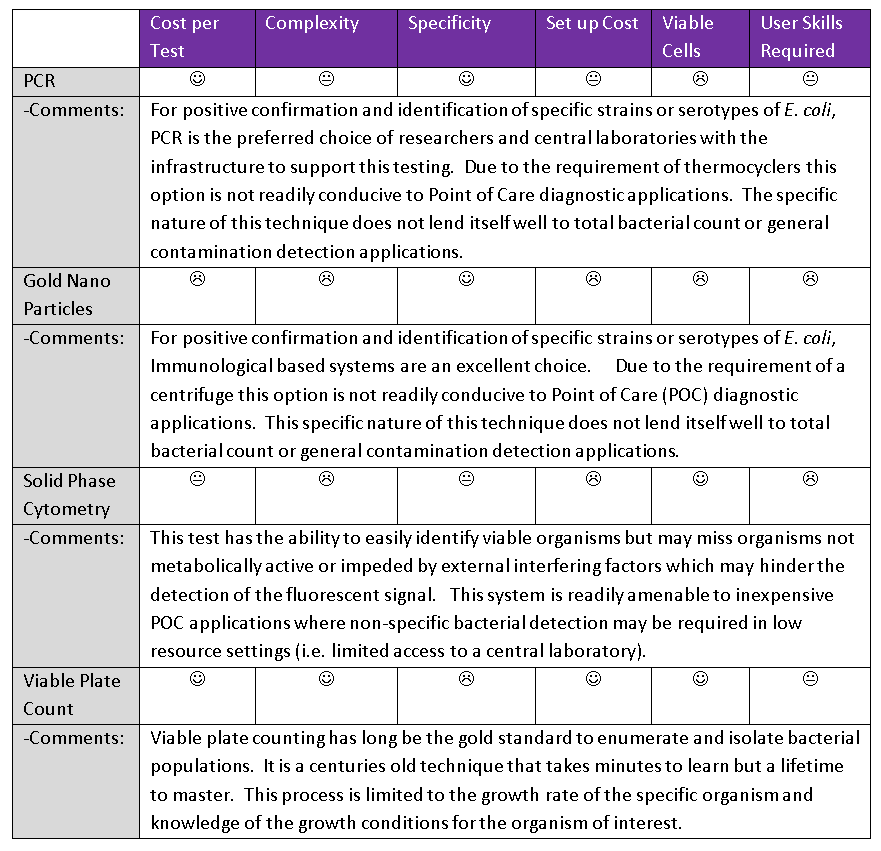
Various methods exist to detect E. coli, amongst them are PCR, gold nanoparticles for a visual colour change confirmation and fluorescent labelled enzymes. This article outlines the detection methods mentioned above, and helps narrow down which detection method is best suited to detect E. coli for a specific application.
PCR/Polymerase Chain Reaction
PCR is the process of taking a single copy of a gene, and through various heating and cooling cycles, multiplying the genes for easier detection. The PCR technique was invented in 1985 by Kary B. Mullis, while working as a chemist at Cetus Corporation[1]
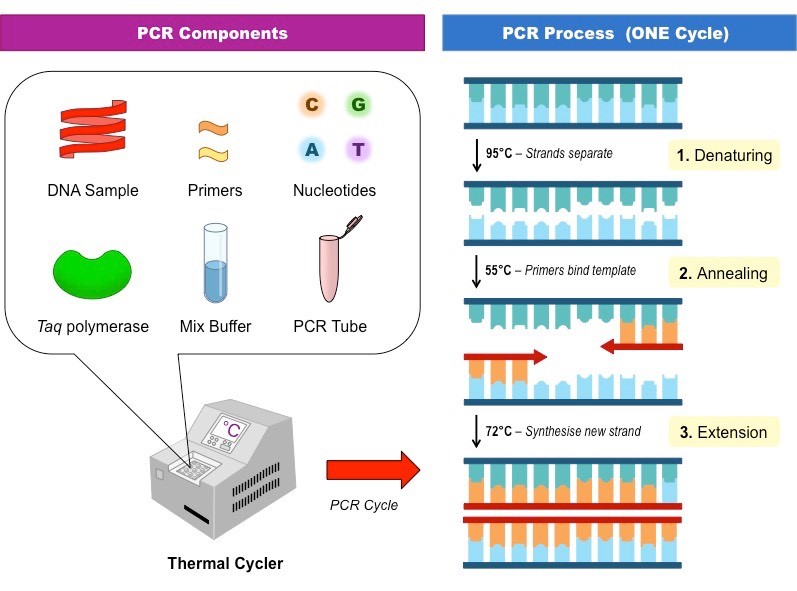
The process of PCR takes place in a thermal cycler due to the various temperatures required to complete a cycle. There are three steps required to complete one cycle of PCR:
- Denaturation- The DNA strand is heated to around 95oC for 1 minute, to separate the two strands.
- Annealing- The temperature drops to around 55oC for 1 minute as DNA primers attach to the 3’ end of the DNA strands.
- Elongation- Taq DNA polymerase bonds to the primer and makes a copy of the strand, the cycle is at around 72oC for 2 minutes. Taq polymerase is a heat resistant enzyme, able to withstand the high temperatures cycles of PCR.
Every ‘cycle’ of PCR doubles the amount of DNA, so 30 cycles of PCR can create roughly 1 billion copies of the DNA strand. Once the large quantity of DNA has been created, labs can isolate and detect the sequence of interest. This is used in many places for rapid disease detection, such as water contamination for E. coli. Due to the specificity of this system, PCR is a powerful tool to help screen and detect E. coli of interest (such as O157:H7) from the relatively harmless environmental strains.
The benefits of PCR to detect E. coli are:
- The assay is extremely specific (it relies on unique DNA for typing).
- The assay is extremely sensitive, functional with fragments of DNA.
- Cost per test is in the order of fractions of cent.
The cons of PCR are:
- Requires trained personnel.
- Knowledge of the specific sequence of target DNA.
- Very specific to targeted coli only.
- Expensive initial capital investment.
Gold Nanoparticles
Exposure to E. coli serotype O157:H7 (even trace amounts in your food) can lead to disease or even death. Gold nanoparticles (Au NPs) can be used for the detection of E. coli, through inductively coupled plasma mass spectrometry (ICPMS). ICPMS is used for detection for trace elemental analysis, being able to rapidly analyze data. [3] In short, a high specific monoclonal antibody against E. coli O157:H7 is bound to the target analyte (the bacteria of interest) and incubated. The solution is then centrifuged to separate the unbound from the bound Au NPs. Upon removal of the supernatant, the remaining cell pellet is digested with an acid solution and the solution is analyzed through ICPMS.
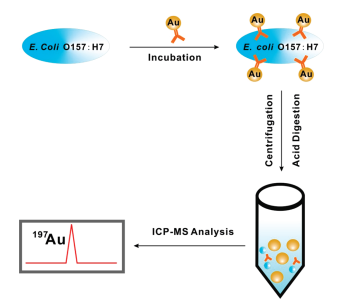
The signal intensity (CPS) vs CFU/mL is plotted; an increasing trend is observed with an increase in bacterial concentration.
The benefits of gold nanoparticles to detect E. coli are:
- The assay is very specific (it relies on unique monoclonal antibodies raised against target antigens usually expressed on the surface of the organism).
- The assay is very sensitive.
- Cost per test is in the order of cents per test.
The cons of gold nanoparticles are:
- Knowledge of the specific sequence of target antigen
- Very specific to targeted antigens
- Requires good conformation and presentation of the target antigens
- Expensive initial capital investment.
- Requires trained personnel.
Solid Phase Cytometry
- coli can also be detected through the use of solid phase cytometry (SPC) in conjunction with fluorescent viability staining. The E. coli in water samples are filtered over a polyester membrane 0.4 µm pore size filter under vacuum to capture the bacteria. The retained E. coli on the filter is then treated with reagents to induce the enzyme β-D-glucuronidase and incubated at 37⁰C for 3 hours. The induced cells are then labelled. Only the enzymes on the viable cells can be labelled, causing the bacteria to cleave the non-fluorescent substrate, leaving only the fluorescent end inside the viable cells.
The fluorescence is then read using the ScanRDI® device, a laser scanning device that enumerates cells via fluorescent labelling. Enumeration of fluorescent cells take up to three minutes. Using this method allows for rapid and visual detection of contaminated water. [5] This system has the advantage of detecting viable and potentially non-culturable cells. Simple biochemical tests such as Biomerieux API strips or more complex systems such as Matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF) performed on isolated pure colonies are further required to positively identify E. coli from other organisms using this technique.
The benefits of Solid Phase Cytometry to detect E. coli are:
- The assay can detect viable organisms
- The assay is limited to the ability to detect the fluorescent signal
- Cost per test is in the order of cents per test.
The cons of Solid Phase Cytometry are:
- The assay can only detect viable organisms
- The assay is limited to the ability to detect the fluorescent signal
- The assay consists of a multi-step process.
- Additional steps are required for confirmation of coli.
Viable cell count
There are many ways to enumerate bacterial concentrations, one simple way is through viable cell counting on an agar plate. The bacterial sample is serially diluted (1:10, 1:100, 1:1000 etc.), in sterile water or Phosphate Buffer Saline (PBS), and plated on Tryptic Soy Agar plates or semi-selective media (such as MacConkey agar to selectively isolate Gram-negative enteric organism). The plates are sealed and incubated in a 37˚C incubator overnight. When counting the colonies the next day, a count between 20 and 200 provides an accurate representation of concentration. [6] As with SPC, Simple biochemical tests such or MALDI-TOF is performed on isolated pure colonies are further required to positively identify E. coli from other organisms using this technique.
The benefits of Solid Phase Cytometry to detect E. coli are:
- The assay can detect viable organisms
- The assay is limited to the ability to detect the culturable organisms
- Cost per test is in the order of cents per test.
The cons of Solid Phase Cytometry are:
- The assay can only detect viable organisms
- The assay is limited to the ability to detect the organisms that grow on your chosen media
- The assay consists of a multi-step process.
- Additional steps are required for confirmation of coli.
Summary
While there are many ways of getting the same results, selecting a method for developing an assay, whether it is for detecting bacteria or simulating fluid, the approach all depends on the application and your target market.
[1] https://en.wikipedia.org/wiki/Kary_Mullis#Accreditation_of_the_PCR_technique
[2] http://ib.bioninja.com.au/standard-level/topic-3-genetics/35-genetic-modification-and/pcr.html
[3]https://www.researchgate.net/publication/42388104_Detection_of_Escherichia_coli_O157H7
[4] https://link.springer.com/content/pdf/10.1007/s00253-009-1879-x.pdf
[5] https://www.ncbi.nlm.nih.gov/pubmed/11777581
[6] https://courses.lumenlearning.com/boundless-microbiology/chapter/counting-bacteria/
Azra Rajwani is a StarFish Medical Biomedical Engineering Lab Technician. She is a recent graduate of the UVIC Biomedical Mechanical Engineering Program, was a former co-op student at StarFish Medical, and this is her first StarFish blog. Some of Azra’s prior projects include a water quality project at UVic; and working in the Neuroscience Department at U of Calgary to better understand neural networks.

