Why a CoVID-19 Point of Care Diagnostic will help insure our future
CoVID-19 Point of Care Diagnostic is important
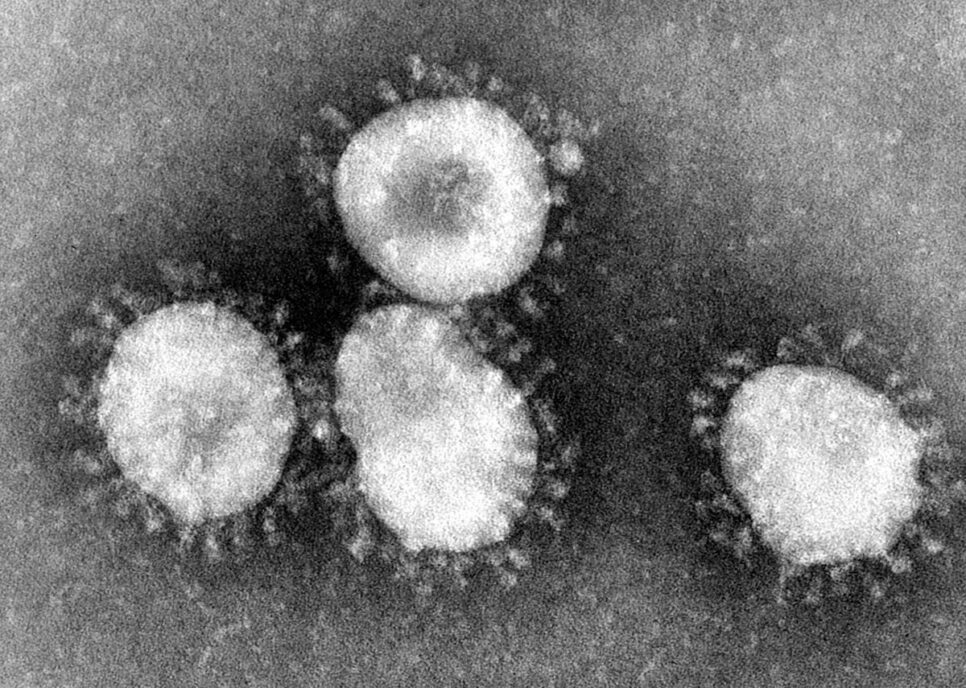
The virus that causes CoViD19 is a coronavirus. The viruses are so called because the virus physically looks like a crown (‘corona’ derived from ancient Greek word for crown) when imaged by electron microscopy. These coronaviruses are a group of enveloped single stranded RNA viruses which typically are responsible for the common cold. About seven kinds are known to cause an infection in humans. Four of these are seasonal and cause the common cold year after year after year. The remaining three coronaviruses are termed emergent coronaviruses because they have been the cause of outbreaks and epidemics. That is why a CoVID-19 Point of Care Diagnostic is so important.
The first emergent coronavirus was the Severe Acute Respiratory Syndrome (SARS) in 2002. Ten years later we had Middle East Respiratory Syndrome (MERS) and now we have a third human coronavirus: Severe Acute Respiratory Syndrome Coronavirus 2 or SARS-CoV-2, which is official name for the virus which causes CoVID-19 (Corona Virus Disease 2019) – so named by the world health organization to make it easy to pronounce globally and to limit geographic specificity.
What makes CoVID-19 unique?
The CoVID-19 virus is a relative of the SARS virus. It’s approximately 80% identical to SARS with a few key differences:
- Preliminary data suggests that this virus behaves more like influenza with the viral load peaking earlier. In other words, when a patient who is infected or when an individual infected first develops symptoms, that’s when the amount of virus is at its highest. When a patient appears to get worse and must be admitted to the hospital, for instance, that’s actually when the viral titer goes down. This suggests that this virus fundamentally has some different transmissibility properties than SARS and may explain why we see an increased transmissibility. This is why it is critically important to follow good social distancing practices and follow self-isolation guidelines.
- The economic impact of the anticipated global pandemic is projected to cost trillions of dollars in economic losses (opposed to the paltry $40 billion caused by SARS).
- Technological advances and new diagnostic capabilities available in 2020 (nearly two decades since SARS) makes the development of infectious disease diagnostics an entirely different enterprise. The advent of sequencing technology that we call genomic technologies have changed the way we approach outbreaks in a significant way. To put this into perspective, it took weeks to even identify SARS as a novel virus. First, we had to grow the SARS virus. Then we had to use other technologies such as micro array technologies to diagnose the infection. CoVID-19 was different. With CoVID-19, the actual sequence of the virus was available as soon as a week after it was identified. Immediately upon identification, Chinese scientists were able to sequence the genome, the genetic makeup of this particular virus. This is critically important because it’s essentially the genome sequence that allows you to develop tests to diagnose an infection that can detect the virus. Having the sequence allows you to develop antibody tests for doing surveillance, as well as potential therapies and vaccines.
CoVID-19 Point of Care Diagnostic Tests
Currently all CoVID-19 Point of Care diagnostic tests being performed so far are primarily using commercially available PCR kits. In December, the Chinese CDC very quickly sequenced and reported the genomic sequence of SARS-CoV-2. This led to the rapid development of PCR kits by a number of companies. There are already eleven commercial kits approved by the FDA, with more than 100 products under registration now.
The issue, however, is that these kits are being developed in a very short time which is an anomaly for the normal timelines required to develop a PCR test. In particular, clinical validation using real world samples is a requirement and this typically would take a couple of years to complete. Given the demand for tests, these kits are validated using analytical sensitivity rather than the more relevant clinical sensitivity.
Sampling and test methods
Another issue is sampling and test methods. Typically, the sampling is done using the nasal or throat swab or oral pharyngeal swabs. These samples are relatively messy comparing to a blood draw. These swabs have a lot of impurities. Also, the skills of people taking these samples impact the sample. Finally, as SARS-CoV-2 is a RNA virus, preservation, storage and transportation of samples is of critical importance as the viral RNA is very unstable.
At the end of the diagnostic chain, the individual laboratory practices where the samples are processed, and the final diagnosis is made is also a critical step. Adding to the complexity is the very limited amount of clinical validation of the non-standardized PCR kits from various multiple commercial sources. Taken together, any of the issues in these steps could contribute to the false negative for the ability to detect the virus infections.
Clinical sensitivity of the test is particularly challenging because this is a new virus – much of the data with regards to the viral load and how much virus is actually present in the nasal swab when you collect it from a patient was actually only published recently.[1]
Best time to do the testing
All this leads to the issue of when is the best time to do the testing? Do we do the testing before a patient gets symptoms? Do we do testing once the patient becomes symptomatic? Does this happen once the patient becomes symptomatic enough that he or she needs to be admitted to the hospital to receive care? Or is it when there is more intensive care in the hospital? Or do we do continue testing after a patient recovers?
With SARS, it was easiest to diagnose the infection when the patients were extremely ill and in the intensive care unit. CoVID-19 appears to be different. A person with the virus can be asymptomatic, have no symptoms, and yet actually infect someone else. In addition, a person could be infected with the virus and never have symptoms (asymptomatic infection). At this point, they might still be contagious. This is why testing for the virus is extremely challenging. The actual amounts of the virus that we are detecting with these tests don’t always correlate with the symptoms, or whether a patient has symptoms at all, or the severity of his or her symptoms.
In other words, CoVID-19 may be easiest to detect when the symptoms are just beginning (or perhaps even before they present), and it may be difficult to get a positive result when very ill patients come in. Additionally, it appears that the viral titers fluctuate over time. As a result, a person could have a positive PCR test one day, the next day negative, and the day after a positive result. This makes it very difficult to use direct detection methods like PCR to confidently characterize or diagnosis infection.
One solution is to develop an assay that uses indirect methods.
A method that is commonly used in infectious diseases diagnostics is to look for antibodies to the virus in the patient. For example, if you are infected with measles or if have been vaccinated for it, your body will generate antibodies to the measles virus. Similarly, infection with the CoVID virus leads your body to generate antibodies. Those antibodies can be used to detect infection or to detect the fact that you were exposed to the virus. Unfortunately, this is not very useful in the acute setting because it usually takes a week or two for your body to make the antibodies.
Host response
Another approach is that potentially we can diagnose the infection by looking at your host response. This is where some of the newer technologies such as RNA sequencing may come to play. With RNA sequencing the patient’s host’s response to this virus can be detected. The idea is that the host response can be specific to the kind of infection. This could mean potentially diagnosing an infection like CoVID-19 without even being able to detect the virus. You’re simply diagnosing it based on the patient’s host response or immune response to that infection. This technology has been demonstrated for influenza and there are clinical tests being developed based on those responses.
Some of these host response markers may also prove to be helpful in figuring out which patients are going to be the sickest and shed some light on the 2.3% mortality issue with CoVID-19.
Prognostic capability
Diagnostic assays based on this technology may have prognostic capability. The idea is that by looking at host response markers on a patient that enters a hospital you might be able to predict who is going to get worse and who is going to get better. This is going to be useful information that could bolster our ability to predict disease outcomes.
In a global pandemic, this capability would greatly help responders prioritize cases and determine who should be kept in quarantine, who should get ICU treatment, who could be safely discharged to self-quarantine at home, etc. Unfortunately, it is likely that very few countries in the world are prepared to receive an influx of hundreds and thousands of cases (this is why it is imperative that we all do our part and practice social distancing). An infectious disease diagnostic with this capability would be an important tool to reduce the burden on the taxed infrastructure in these situations.
Get a test in the mail
Another way of avoiding this is developing a test that would not require a visit to the hospital at all. Quarantine is a powerful tool in the prevention of global pandemics. A better approach to queuing up at a hospital while wearing a mask to get tested would be to stay at home and get a test in the mail from the doctor. This would eliminate queueing up with a group of sick people.
Researchers at the Broad Institute and MIT McGovern Institute have been developing a diagnostics method based on CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) proteins to be able to program a CRISPR protein to recognize unique signatures that are found in different kinds of viral or bacterial pathogens.[2]
SHERLOCK
One of the techniques that these researchers are working on is called SHERLOCK.[3] The idea of SHERLOCK is that one day there will be a paper strip-based test where a patient can provide either blood, urine or saliva sample. By mixing the sample with the CRISPER proteins and a short RNA that’s been designed to recognize the unique signature on these viral sequences, the protein/RNA pairs will carry out a reaction that allows these papers to be able to tell you in a very simple way (like a pregnancy test) whether or not infection is present. At the time of preparing this article, the institute has not yet had access to real patient samples and preliminary development has only been working with artificial chemically synthesized fragments of those viruses. However, this process has been shown to have promise in the ability to detect these viral sequences at fairly low concentration.
This platform is very exciting because the amount of time that it takes to redesign or redevelop this test to address a new infectious pathogen can be a very quick turnaround. Currently CoVID-19 is the focus of a great amount of research in infectious disease point of care diagnostics development, but in the future hopefully all the things that we learn from this process can be applied to other diseases as well.
What is needed now
What is needed now is a reliable and a rapid test for people to be on the point of care basis or home-based tests. Especially now as China returns to work and a lot of the employers and government agencies want their employees tested before they go back to work. Testing millions of people through a PCR test is a daunting task. The turnaround time is typically about two to three hours for a PCR test.
More importantly, if you’re testing hundreds of thousands of people, how do you process that many people into a safe area for sampling? This congregation for testing creates opportunities for cross infection and is defeating the purpose of quarantine. The need for a rapid test as well as self-administered sampling is very important.
CoVID-19 is a global wakeup call 20 years after SARS. We must be vigilant and continue to develop point of care infectious disease diagnostics to prepare for the next global pandemic. With only a 2.3% mortality (for reference regular seasonal influenza has a mortality of 0.1% or ~200,000 annually[4]), we got off relatively lucky. It is not a matter of if, but when. We need to be prepared. Continued investments in new Point of Care Infectious Disease Assays and platforms like a CoVID-19 Point of Care Diagnostic will help insure our collective futures for the next global pandemic.
This blog was prepared based on the proceedings of the WUXI online forum on COVID-19, summarizing the thoughts and interviews of Nick Naclerio, Charles Chiu, Feng Zhang and Victor Shi. For more information on this topic please see: https://www.broadinstitute.org/news/enabling-coronavirus-detection-using-crispr-cas13-open-access-sherlock-research-protocol
[1] Zou et al. 2020. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. NEJM. Feb 20, 2020. DOI: 10.1056/NEJMc2001737
[2] https://www.broadinstitute.org/news/enabling-coronavirus-detection-using-crispr-cas13-open-access-sherlock-research-protocol
[3] https://www.broadinstitute.org/news/scientists-unveil-crispr-based-diagnostic-platform
[4] GBD 2017 Influenza Collaborators Mortality, morbidity, and hospitalizations due to influenza lower respiratory tract infections, 2017: an analysis for the Global Burden of Disease Study 2017. Lancet Respir Med. 2019;7:69-89. 10.1016/S2213-2600(18)30496-X
Nick Allan is the StarFish Medical Bio Services Manager. He applies creative thinking and innovation to biomedical project commercialization from product definition through sustaining engineering.
Photo Credit: Content Providers(s): CDC/Dr. Fred Murphy – This media comes from the Centers for Disease Control and Prevention‘s Public Health Image Library (PHIL), with identification number #4814.
Read how StarFish Medical led a consortium that created a Ventilator 2.0 therapy device in record time.
